Huang reparameterized Gompertz survival model
Source:R/huang_reparam_gompertz_survival.R
HuangRGS.RdHuangRGS reparametrized Gompertz survival model for microbial inactivation.
Returns the model parameters estimated according to data collected in microbial inactivation experiments.
Arguments
- x
is a numeric vector indicating the heating time under a constant temperature of the experiment
- Y0
is the initial microbial concentration (log10(cfu 1/g))
- k
is the inactivation rate (1/s)
- M
is a time constant (s)
Details
The model's inputs are:
t: time, assuming time zero as the beginning of the experiment.
Y(t): the base 10 logarithm of the bacterial concentration ($log10(X(t)$) measured at time t.
Users should make sure that the bacterial concentration input is entered
in base 10 logarithm, Y(t) = log10(X(t)).
References
Huang L (2009). “Thermal inactivation of Listeria monocytogenes in ground beef under isothermal and dynamic temperature conditions.” Journal of Food Engineering, 90(3), 380-387. ISSN 0260-8774, doi:10.1016/j.jfoodeng.2008.07.011 , https://www.sciencedirect.com/science/article/pii/S0260877408003439.
Author
Vasco Cadavez vcadavez@ipb.pt and Ursula Gonzales-Barron ubarron@ipb.pt
Examples
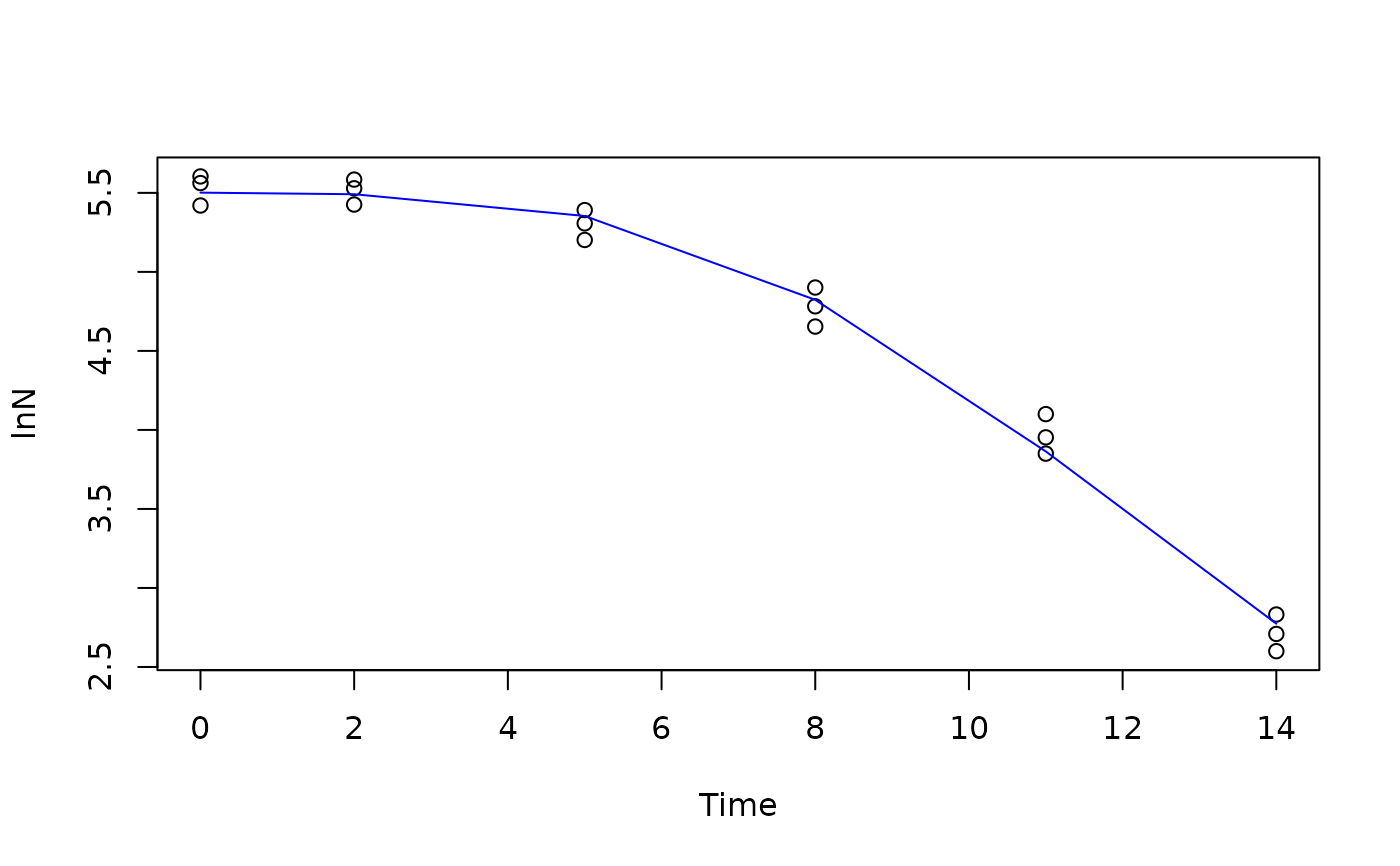
library(gslnls)
data(bixina)
initial_values = list(Y0=5.6, k=0.37, M=6.8)
fit <- gsl_nls(lnN ~ HuangRGS(Time, Y0, k, M),
data=bixina,
start = initial_values)
summary(fit)
#>
#> Formula: lnN ~ HuangRGS(Time, Y0, k, M)
#>
#> Parameters:
#> Estimate Std. Error t value Pr(>|t|)
#> Y0 5.502090 0.046071 119.43 < 2e-16 ***
#> k 0.182138 0.009672 18.83 7.53e-12 ***
#> M 12.053884 0.150299 80.20 < 2e-16 ***
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> Residual standard error: 0.1154 on 15 degrees of freedom
#>
#> Number of iterations to convergence: 12
#> Achieved convergence tolerance: 1.786e-12
#>
plot(lnN ~ Time, data=bixina)
lines(bixina$Time, predict(fit), col="blue")