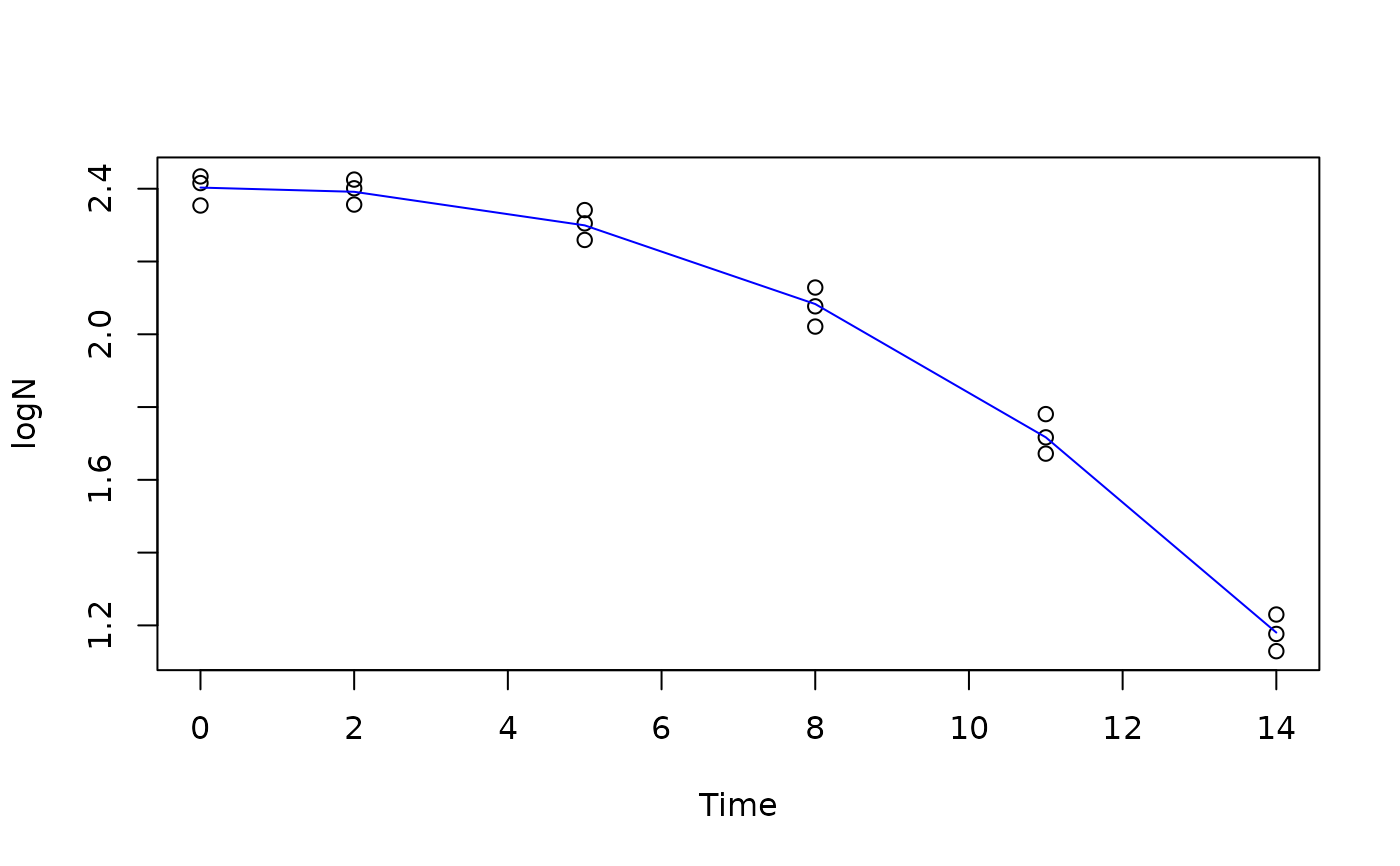
WeibullM inactivation model for microbial inactivation curve.
Returns the model parameters estimated according to data collected in microbial inactivation experiments.
Arguments
- x
is a numeric vector indicating the heating time under a constant temperature of the experiment
- Y0
is the base 10 logarithm of the initial (time=0) bacterial concentration (N0)
- sigma
is the time of first decimal reduction
- alpha
which is a shape parameter
Details
The model's inputs are:
t: time, assuming time zero as the beginning of the experiment.
Y(t): the base 10 logarithm of the bacterial concentration ($log10(N(t)$) measured at time t.
Users should make sure that the bacterial concentration input is entered
in base 10 logarithm, Y(t) = log10(N(t)).
References
Mafart P, Couvert O, Gaillard S, Leguerinel (2002). “On calculating sterility in thermal preservation methods: application of the Weibull frequency distribution model.” International Journal of Food Microbiology, 72, 107-113.
Author
Vasco Cadavez vcadavez@ipb.pt and Ursula Gonzales-Barron ubarron@ipb.pt
Examples
library(gslnls)
data(bixina)
initial_values = list(Y0=2.5, sigma=2, alpha=2)
bixina$N <- exp(bixina$lnN)
bixina$logN <- log10(bixina$N)
fit <- gsl_nls(logN ~ WeibullM(Time,Y0,sigma, alpha),
data=bixina,
start = initial_values)
summary(fit)
#>
#> Formula: logN ~ WeibullM(Time, Y0, sigma, alpha)
#>
#> Parameters:
#> Estimate Std. Error t value Pr(>|t|)
#> Y0 2.40313 0.01691 142.09 < 2e-16 ***
#> sigma 12.87224 0.13543 95.05 < 2e-16 ***
#> alpha 2.39534 0.13162 18.20 1.23e-11 ***
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> Residual standard error: 0.042 on 15 degrees of freedom
#>
#> Number of iterations to convergence: 14
#> Achieved convergence tolerance: 1.208e-08
#>
plot(logN ~ Time, data=bixina)
lines(bixina$Time, predict(fit), col="blue")